Poincare map and return time - interval version
Computing rigorous bounds for Poincare map is quite similar to nonrigorous version - see Poincare maps - nonrigorous methods. We have to specify initial condition as doubleton set (see Representation of initial condition)
and call operator of class IPoincareMap
The following example is a complete proof of the existence of Lyapunov orbit L1 in the Planar Restricted Circular Three Body Problem for a fixed Jacobi constant.
Complete example (from examples/poincare/IPoincareMapExample.cpp):
Derivative of Poincare map.
The class IPoincareMap provides an overloaded operator for computation of Poincare map and its derivative. First, an initial condition for the main equation and for the variational equation must be specified - see Representation of initial condition.
We have to define an interval matrix that will store bound for set of monodromy matrices 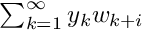 where
where 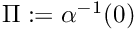 and
and 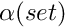 is a bound for the return time.
is a bound for the return time.
Eventually we have to call an operator
- Note
- The last argument
returnTimeis optional and can be skipped for autonomous systems
After successful integration we have
Pbound for the Poincare map at the set of initial conditionsset- bound for the monodromy matrix
Given monodromy matrix we can recompute it to derivative of Poincare map by
- Note
- for nonautonomous systems the argument
returnTimecan be skipped
- Note
- we split computation of derivative of Poincare map into computation of monodromy matrix and solving implicit equation. This is because the user can provide own and optimized routine for recomputation of monodromy matrix to derivative of Poincare map. For user convenience we provide a general routine
computeDP.
- Note
- The matrix
DPreturned bycomputeDPis in full dimension. One can take a submatrix ofDPthat correspond to variables on the section.
Below we give an easy example of computing derivative of Poincare map. See also more advanced examples
- proof of the existence od 3 different periodic orbits for the Rossler system Periodic orbits for the Rossler system
- proof of the existence of 116 periodic orbits for the Lorenz system Multiple periodic solutions for the Lorenz system
Complete example (from examples/poincare/IPoincareMapDerivativeExample.cpp):
